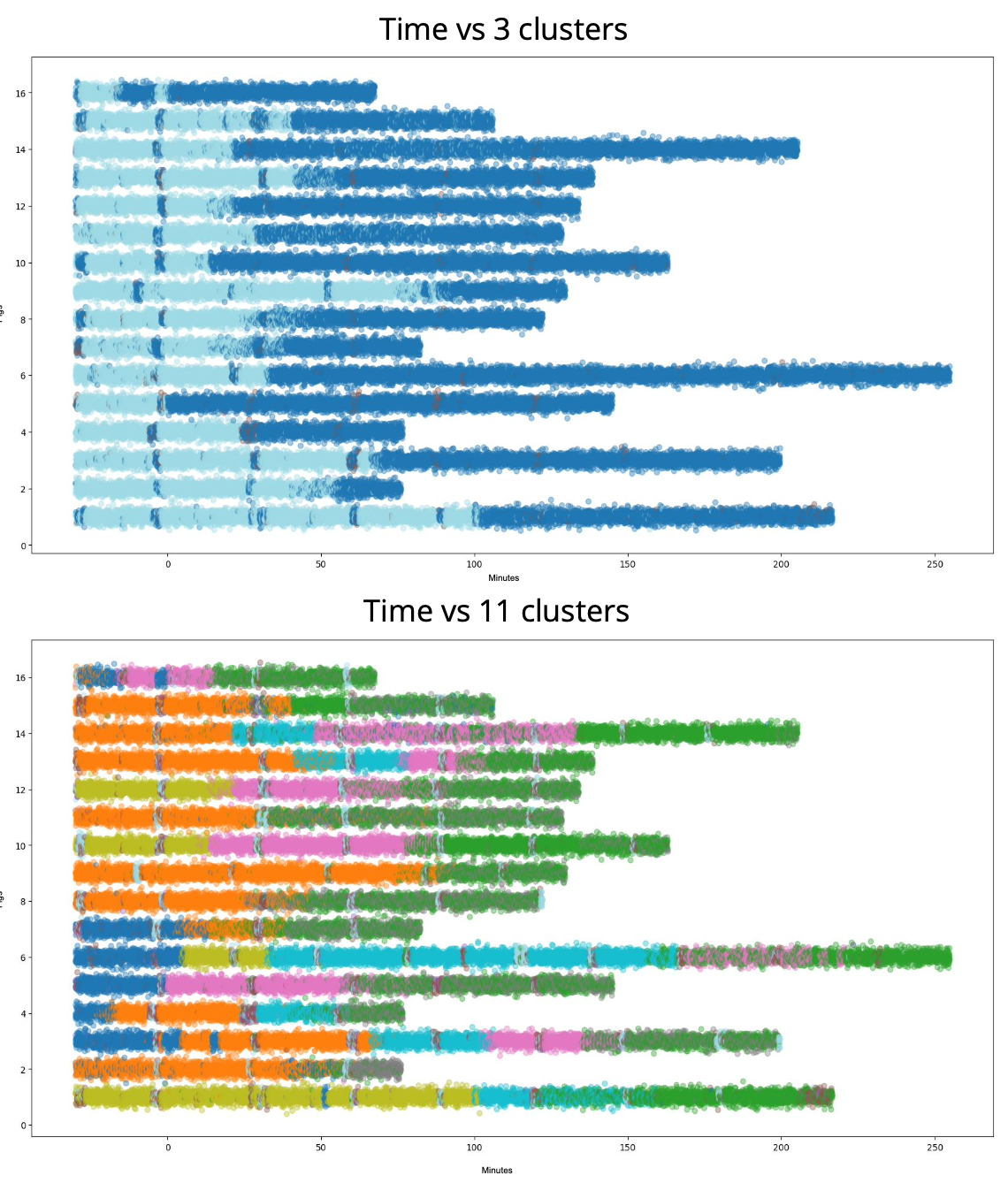
Physiologic state clustering in shock
While we often strive to build intuitive and interpretable models based on data features we deem meaningful and potentially related to the phenomenon we aim to understand, this approach inherently limits the potential for the learned models to make novel discoveries and uncover previously unknown relationships. I had the privilege of mentoring Chufan Gao during his time as an undergraduate researcher in CMU’s Robotics Institute Summer Scholars (RISS) program. Building upon our investigations into modeling and predicting hemodynamic shock (you can read more about that project here), Chufan analyzed the same data through a more unbiased lens, employing various deep neural network architectures to directly encode and model the raw waveform data. His poster is available in the gallery, and I encourage you to explore this work further in his NeurIPS abstract (Gao et al., 2019).